Appraising the causal relationship between plasma caffeine levels and neuropsychiatric disorders through Mendelian ... - BMC Medicine
Study overview
This study applied a two-sample MR design with publicly available summary-level data taken from meta-analyses of genome-wide association studies (GWASs) as well as the FinnGen study, which was not included in any of the GWAS meta-analyses (Fig. 1). Outcomes included in this study were anorexia nervosa, bipolar disorder, MDD, and schizophrenia. Brief information about the used data sources for plasma caffeine and outcomes are described below. More comprehensive details for each GWAS are described in the corresponding articles [9,10,11,12,13,14].
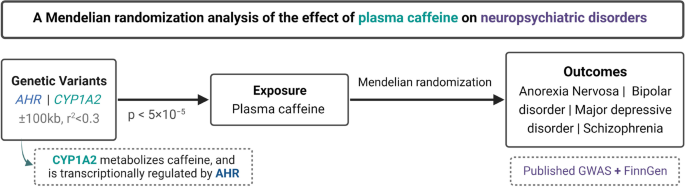
Study design overview (created with BioRender.com). AHR, aryl hydrocarbon receptor; CYP1A2, cytochrome P450 1A2 (CYP1A2); GWAS, genome-wide association study
Data sources
Plasma caffeine GWAS
Cornelis et al. performed a meta-analysis of summary statistics from six plasma caffeine (1,3,7-trimethylxanthine) GWASs comprising 9876 participants of mainly European ancestry, with mean ages between 47 and 71 years [9, 15]. Participants were asked to fast prior to measurements being taken, and each study standardized plasma caffeine measurements. Association estimates accounted for confounding by smoking status and, when applicable, age, sex, and principal components of ancestry.
Anorexia nervosa GWAS
Watson et al.'s GWAS was a meta-analysis from the Psychiatric Genetic Consortium (PGC) Eating Disorders Working Group, which contained 16,992 cases of anorexia nervosa and 55,525 controls from 33 studies—mostly of European ancestry individuals [14, 16]. Studies were asked to adjust their analyses for the first five principal components of ancestry. The linkage disequilibrium score regression intercept for the GWAS implied the presence of an insignificant residual population structure.
Bipolar disorder GWAS
This study is a meta-analysis of 41,917 cases of bipolar disorder and 371,549 controls from the PGC and five population-based cohort studies including the UK Biobank (UKB) [16, 17]. Samples were restricted to participants of European ancestry, and cases were diagnosed using the Diagnostic and Statistics Manual or International Classification of Diseases guidelines. The genomic control inflation factor was between 0.97 and 1.05 for the participating studies, implying minimal residual population structure.
MDD GWAS
Howard et al. meta-analyzed GWAS summary statistics of 170,756 MDD cases and 329,443 controls from the PGC and the UK Biobank [11]. All study samples largely comprised individuals of European ancestry. Cases were identified using a combination of self-report and medical records. The linkage disequilibrium (LD) score regression intercept for the GWAS summary statistics used is 1.02 (standard error = 0.01) implying the presence of insignificant residual population structure. GWAS summary statistics were extracted from the OpenGWAS platform using the ID: ieu-b-102 [18, 19].
Schizophrenia GWAS
Trubetskoy et al. meta-analyzed genetic data from 90 cohorts within the PGC [10, 16]. The resulting sample included 76,755 schizophrenia cases and 243,649 controls. Participating studies used genomic quality control to control for inflated test statistics and adjusted for at least four principal components of ancestry.
FinnGen study
The FinnGen study is a large (n = 356,077 in round 8) population biobank, based in Finland, described in detail elsewhere [20, 21]. GWAS summary data on the FinnGen cohort (round 8) includes 390 individuals with anorexia nervosa (FinnGen phenotype ID: R18 ANOREXIA), 6562 with bipolar disorder (F5 BIPO), 39,747 with MDD (F5 DEPRESSIO), and 6522 with schizophrenia (F5 SCHZPHR) [22]. Cases were identified from medical records [20, 21]. These GWASs adjusted for age, sex, the first 10 genetic principal components, and genotyping batch [12].
Mendelian randomization analysis
We selected single-nucleotide polymorphisms (SNPs) that were strongly associated (p < 5 × 10−5) with plasma caffeine levels and located within 100 kb of the CYP1A2 and aryl hydrocarbon receptor (AHR) gene regions as instrumental variables (GRCh37/hg19 assembly by Ensembl: 15:75041185-75048543 and 7:17338246-17385776, respectively). These genes were selected owing to their role in caffeine metabolism [9]. Variants in these gene regions have been used as instrumental variables for plasma caffeine in previous MR studies [23, 24]. The statistical significance threshold of p < 5 × 10−5 was selected to ensure that the used SNPs would be strong instruments for plasma caffeine, while accounting for gene region-wide (a Bonferroni correction for the 955 SNPs measured by Cornelis et al. within the two gene regions provides a p ~ < 5 × 10−5). SNPs were clumped with a r2 of 0.3 and 10,000 kb windows using the TwoSampleMR R package [25, 26]. We used the false discovery rate inverse quantile transformation (FIQT) Winner's curse correction to account for Winner's curse [27]. We harmonized data sources using TwoSampleMR and excluded palindromic SNPs, which could not be aligned based on their allele frequency.
We firstly meta-analyzed the SNP-outcome associations from each GWAS meta-analysis with the equivalent outcome data in the FinnGen study using an inverse variance weighted meta-analysis. Secondly, we conducted the MR analysis. The primary estimator was the Wald ratio, which was defined as the ratio of the SNP-outcome association to the SNP-exposure association. Wald ratios were combined using a multiplicative inverse variance-weighted (IVW) random effects model, using SNPs' LD matrix estimated from the European subsample of the 1000 Genomes Project to account for correlations between variants [28]. This was implemented using the code for random effects IVW (accounting for correlations) by Burgess et al. [28]. A correlated variants IVW estimator was chosen over a more conventional (independent variant) IVW estimator because of the potential to improve the precision of MR estimates in a cis setting. We used the Benjamini–Hochberg procedure to correct for multiple testing. A false discovery rate (FDR) adjusted p-value < 0.05 was regarded as statistically significant. Odds ratios (OR) presented here represent the multiplicative increase in the odds for each standard deviation (SD) increase in plasma caffeine levels.
Heterogeneity tests have been proposed as sensitivity analyses for pleiotropy in MR studies [29]. Because these methods have not been extended to cis settings with correlated variants, we first explore the robustness of outlier SNPs using leave-one-out analyses and then explore heterogeneity between the AHR and CYP1A2 genes. We additionally explore the heterogeneity in the MR estimates between data sources and replicate our analysis using only the lead SNP from each gene region (rs4410790 for AHR and rs2472297 for CYP1A2). To the best of our knowledge, there is no sample overlap between the exposure and outcome GWASs.
Comments
Post a Comment